Abstract
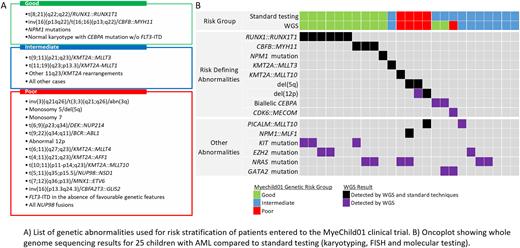
Background Genetics is important in the diagnosis of paediatric acute myeloid leukaemia (AML). A number of chromosomal abnormalities and mutations have prognostic significance and timely genetic analysis is essential for risk stratification. The recent Myechild 01 clinical trial for children with AML required testing for >20 different genetic abnormalities to assign newly diagnosed patients into three prognostic genetic risk groups: good, intermediate and poor (Figure A). Throughout the trial a combination of techniques was employed to detect these abnormalities; including chromosomal analysis, FISH for cytogenetically cryptic abnormalities, notably the poor risk fusions: t(5;11)(q35;p15)/NUP98::NSD1, inv(16)(p13q24)/CBFA2T3::GLIS2, t(7;12)(q36;p13)/ETV6::MNX1, as well as molecular testing for NPM1, FLT3-ITD and CEBPA mutations. Here, we present a small retrospective study demonstrating that whole genome sequencing (WGS) has the potential to detect all prognostically significant abnormalities in childhood AML within a single test.
Methods WGS was performed on matched diagnostic and remission samples from 25 paediatric AML patients entered to the Myechild 01 treatment trial. Remission samples, minimal residual disease negative by flow cytometry and/or molecular testing, were used as germline controls. For validation, whole transcriptome sequencing (WTS) was performed on diagnostic RNA from each patient in the study.
Results Leukaemia associated genetic abnormalities were detected in all 25 patients, including structural variants associated with gene fusions (n=14), single nucleotide variants (SNV) (n=20) and/or copy number abnormalities/aneuploidies (n=17). All gene fusions and mutations were confirmed by WTS.
WGS reliably detected all risk stratifying abnormalities identified by standard techniques (Figure B), including 11 patients with gene fusions: RUNX1::RUNX1T1 (n=6), CBFB::MYH11 (n=3), KMT2A::MLLT3 (n=1) and KMT2A::MLLT10 (n=1) and 3 patients with copy number abnormalities: del(5q) (n=2) and del(12p) (n=1) and one patient with an NPM1 mutation.
Among 10 patients with no risk stratifying abnormalities detected by standard techniques, WGS identified risk defining genetic features in 3 of them. Two patients could be reclassified as good risk due to the discovery of biallelic CEBPA mutations. Interestingly, one of the CEBPA positive patients presented with a homozygous mutation (VAF=0.97), which WGS revealed to be the consequence of copy number neutral loss of heterozygosity for the long arm of chromosome 19.
A single patient could be reclassified as poor risk due to the finding of a rearrangement of MECOM with CDK6. Cytogenetics showed a deletion of 7q as the sole karyotypic abnormality, which was revealed to be an unbalanced der(7)t(3;7)(q21.2;q26.2) translocation by WGS. Over-expression of MECOM was confirmed by WTS. This patient also had a GATA2 mutation.
A risk defining abnormality was not detected by either standard testing or WGS in 7 patients, however 5 of them presented with chromosomal abnormalities which were confirmed and further characterised by WGS, including aneuploidies (n=3) and/or structural rearrangements (n=4).
Across the cohort, WGS detected a wide range of abnormalities not currently used in the risk stratification algorithm, including the fusions genes: NPM1::MLF1 (n=1) and PICALM::MLLT10 (n=2), which, although not stratifying in Myechild 01, have been reported to be associated with a good and a poor prognosis, respectively. SNV in genes commonly mutated in AML were observed in 19 patients (76%) including NRAS (n=6), KIT (n=3) EZH2 (n=3) and GATA2 (n=2). It has been suggested that the co-occurrence of mutations may be used to refine risk groups in childhood AML, notably the presence of KIT and NRAS mutations in RUNX1::RUNXT1 and CBFB::MYH11 positive leukaemia, respectively.
Conclusion This small pilot study has demonstrated the ability of WGS to identify a wide range of genetic abnormalities that are currently used for risk stratification in childhood AML. The full spectrum of genomic events observed here highlights the potential of WGS to refine risk stratification and identify novel genetic changes as therapeutic targets in future trials.
Disclosures
Kingsbury:Illumina: Current Employment, Current equity holder in publicly-traded company. James:Illumina: Current Employment, Current equity holder in publicly-traded company. Mijuskovic:Illumina: Current Employment, Current equity holder in publicly-traded company. Dillon:Abbvie: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Amgen: Research Funding; Astellas: Consultancy, Honoraria, Speakers Bureau; Avencell: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; Jazz: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Novartis: Consultancy, Honoraria, Speakers Bureau; Pfizer: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Shattuck: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees. Ross:Illumina: Current Employment, Current equity holder in publicly-traded company.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal